Ore Mining Analysis
Written on April 14th, 2023 by Frank Hsiung
Data Source
This is an open dataset about ore mining process public on Kaggle community by a single source provider. The contents are basically describing the factors that are used in floating plant to purify the iron ore flow by removing the impurity(silica). For details of the description, visit: Ore mining
Objectives
The main goal is to use this data to predict how much impurity is in the ore concentrate. As this impurity is measured every hour, if we can predict how much silica (impurity) is in the ore concentrate, we can help the engineers, giving them early information to take actions (empowering!). Hence, they will be able to take corrective actions in advance (reduce impurity, if it is the case) and also help the environment (reducing the amount of ore that goes to tailings as you reduce silica in the ore concentrate).
Data workflow
Here’s some of the outlining features I add for this project:
- Memory Saving
- Timeseries Data Integrity
- Exploratory Data Analysis(EDA)
- ML model(LightGBM)
Memory Saving
At first, this notebook was implement and experimented on kaggle for it’s data origin and compatibility of swift testing and play around for ore mining data. Therefore, it is essential to save more memory for larger dataset, one for potential insufficiency of data RAM, another for faster process speed on publish stage on Kaggle.
def reduce_mem_usage(df, verbose=True):
numerics = ['int16', 'int32', 'int64', 'float16', 'float32', 'float64']
start_mem = df.memory_usage().sum() / 1024**2
for col in df.columns:
col_type = df[col].dtypes
if col_type in numerics:
c_min = df[col].min()
c_max = df[col].max()
if str(col_type)[:3] == 'int':
if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:
df[col] = df[col].astype(np.int8)
elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:
df[col] = df[col].astype(np.int16)
elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:
df[col] = df[col].astype(np.int32)
elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:
df[col] = df[col].astype(np.int64)
else:
if c_min > np.finfo(np.float16).min and c_max < np.finfo(np.float16).max:
df[col] = df[col].astype(np.float32)
elif c_min > np.finfo(np.float32).min and c_max < np.finfo(np.float32).max:
df[col] = df[col].astype(np.float32)
else:
df[col] = df[col].astype(np.float64)
end_mem = df.memory_usage().sum() / 1024**2
if verbose: print('Mem. usage decreased to {:5.2f} Mb ({:.1f}% reduction)'.format(end_mem, 100 * (start_mem - end_mem) / start_mem))
return df
df = pd.read_csv('/kaggle/input/quality-prediction-in-a-mining-process/MiningProcess_Flotation_Plant_Database.csv',decimal=",",parse_dates=["date"],infer_datetime_format=True)
df = reduce_mem_usage(df)
Mem. usage decreased to 70.33 Mb (47.9% reduction)
Features outline
df.head()
| date | %_Iron_Feed | %_Silica_Feed | Starch_Flow | Amina_Flow | Ore_Pulp_Flow | Ore_Pulp_pH | Ore_Pulp_Density | Flotation_Column_01_Air_Flow | Flotation_Column_02_Air_Flow | Flotation_Column_03_Air_Flow | Flotation_Column_04_Air_Flow | Flotation_Column_05_Air_Flow | Flotation_Column_06_Air_Flow | Flotation_Column_07_Air_Flow | Flotation_Column_01_Level | Flotation_Column_02_Level | Flotation_Column_03_Level | Flotation_Column_04_Level | Flotation_Column_05_Level | Flotation_Column_06_Level | Flotation_Column_07_Level | %_Iron_Concentrate | %_Silica_Concentrate | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2017-03-10 01:00:00 | 55.200001 | 16.98 | 3019.530029 | 557.434021 | 395.713013 | 10.0664 | 1.74 | 249.214005 | 253.235001 | 250.576004 | 295.096008 | 306.399994 | 250.225006 | 250.884003 | 457.395996 | 432.962006 | 424.954010 | 443.558014 | 502.255005 | 446.369995 | 523.343994 | 66.910004 | 1.31 |
| 1 | 2017-03-10 01:00:00 | 55.200001 | 16.98 | 3024.409912 | 563.965027 | 397.382996 | 10.0672 | 1.74 | 249.718994 | 250.531998 | 250.862000 | 295.096008 | 306.399994 | 250.136993 | 248.994003 | 451.890991 | 429.559998 | 432.938995 | 448.085999 | 496.363007 | 445.921997 | 498.075012 | 66.910004 | 1.31 |
| 2 | 2017-03-10 01:00:00 | 55.200001 | 16.98 | 3043.459961 | 568.054016 | 399.667999 | 10.0680 | 1.74 | 249.740997 | 247.873993 | 250.313004 | 295.096008 | 306.399994 | 251.345001 | 248.070999 | 451.239990 | 468.927002 | 434.609985 | 449.687988 | 484.411011 | 447.825989 | 458.566986 | 66.910004 | 1.31 |
| 3 | 2017-03-10 01:00:00 | 55.200001 | 16.98 | 3047.360107 | 568.664978 | 397.938995 | 10.0689 | 1.74 | 249.917007 | 254.487000 | 250.048996 | 295.096008 | 306.399994 | 250.421997 | 251.147003 | 452.441010 | 458.165009 | 442.864990 | 446.209991 | 471.411011 | 437.690002 | 427.669006 | 66.910004 | 1.31 |
| 4 | 2017-03-10 01:00:00 | 55.200001 | 16.98 | 3033.689941 | 558.166992 | 400.253998 | 10.0697 | 1.74 | 250.203003 | 252.136002 | 249.895004 | 295.096008 | 306.399994 | 249.983002 | 248.927994 | 452.441010 | 452.899994 | 450.523010 | 453.670013 | 462.597992 | 443.682007 | 425.678986 | 66.910004 | 1.31 |
- Goal is to predict % Silica Concentrate
- Silica Concentrate is the impurity in the iron ore which needs to be removed
- The value of modeling on this target value is that it takes at least an hour to measure the impurity
Timeseries Data Integrity
For this task, there’s an obvious need to treat the data as timeseries format for it’s time continuity and the need mentioned in objectives that we would like to predict the trend and future purity based on current state.
# Set datetime column as index
dt_df = df.set_index('date')
# Pick out discontinuity in data
all_hours = pd.Series(data=pd.date_range(start=dt_df.index.min(), end=dt_df.index.max(), freq='H'))
mask = all_hours.isin(dt_df.index.values)
all_hours[~mask]
149 2017-03-16 06:00:00
150 2017-03-16 07:00:00
151 2017-03-16 08:00:00
152 2017-03-16 09:00:00
153 2017-03-16 10:00:00
...
462 2017-03-29 07:00:00
463 2017-03-29 08:00:00
464 2017-03-29 09:00:00
465 2017-03-29 10:00:00
466 2017-03-29 11:00:00
Length: 318, dtype: datetime64[ns]
Therefore, we choose only to reserve the data after 2017-03-29 12:00:00
dt_df = dt_df.loc["2017-03-29 12:00:00":]
The measurement frequency of most rows in the dataset is every 20 seconds according to what the publisher/author of the dataset mentions. This would imply that there should be 180 measurements in each hour if this information is correct.
dt_df.groupby(dt_df.index).count()["%_Silica_Concentrate"].value_counts()
180 3947
179 1
Name: %_Silica_Concentrate, dtype: int64
dt_df.groupby(dt_df.index).count()["%_Silica_Concentrate"][dt_df.groupby(dt_df.index).count()["%_Silica_Concentrate"] < 180]
date
2017-04-10 179
Name: %_Silica_Concentrate, dtype: int64
dt_df.loc['2017-04-10'].groupby(dt_df.loc['2017-04-10'].index).count()['%_Silica_Concentrate']
date
2017-04-10 00:00:00 179
2017-04-10 01:00:00 180
2017-04-10 02:00:00 180
2017-04-10 03:00:00 180
2017-04-10 04:00:00 180
2017-04-10 05:00:00 180
2017-04-10 06:00:00 180
2017-04-10 07:00:00 180
2017-04-10 08:00:00 180
2017-04-10 09:00:00 180
2017-04-10 10:00:00 180
2017-04-10 11:00:00 180
2017-04-10 12:00:00 180
2017-04-10 13:00:00 180
2017-04-10 14:00:00 180
2017-04-10 15:00:00 180
2017-04-10 16:00:00 180
2017-04-10 17:00:00 180
2017-04-10 18:00:00 180
2017-04-10 19:00:00 180
2017-04-10 20:00:00 180
2017-04-10 21:00:00 180
2017-04-10 22:00:00 180
2017-04-10 23:00:00 180
Name: %_Silica_Concentrate, dtype: int64
We replace that missing timestamp with the latest datapoint behind it, which is 2017-04-09 23:00:00
df_before = dt_df.copy().loc[:'2017-04-10 00:00:00']
df_after = dt_df.copy().loc['2017-04-10 01:00:00':]
new_date = pd.to_datetime('2017-04-10 00:00:00')
new_data = pd.DataFrame(df_before[-1:].values, index=[new_date], columns=df_before.columns)
df_before = pd.concat([df_before,new_data],axis=0)
dt_df = pd.concat([df_before, df_after])
dt_df.reset_index(drop=True)
dt_df["duration"] = 20
dt_df.loc[0,"duration"] = 0
dt_df.duration = dt_df.duration.cumsum()
dt_df['Date_with_seconds'] = pd.Timestamp("2017-03-29 12:00:00") + pd.to_timedelta(dt_df['duration'], unit='s')
dt_df = dt_df.set_index("Date_with_seconds")
EDA
Correlation Heatmap
fig = plt.figure(figsize=(18,18))
sns.heatmap(dt_df.corr(), annot=True, cmap='viridis')
<AxesSubplot:>
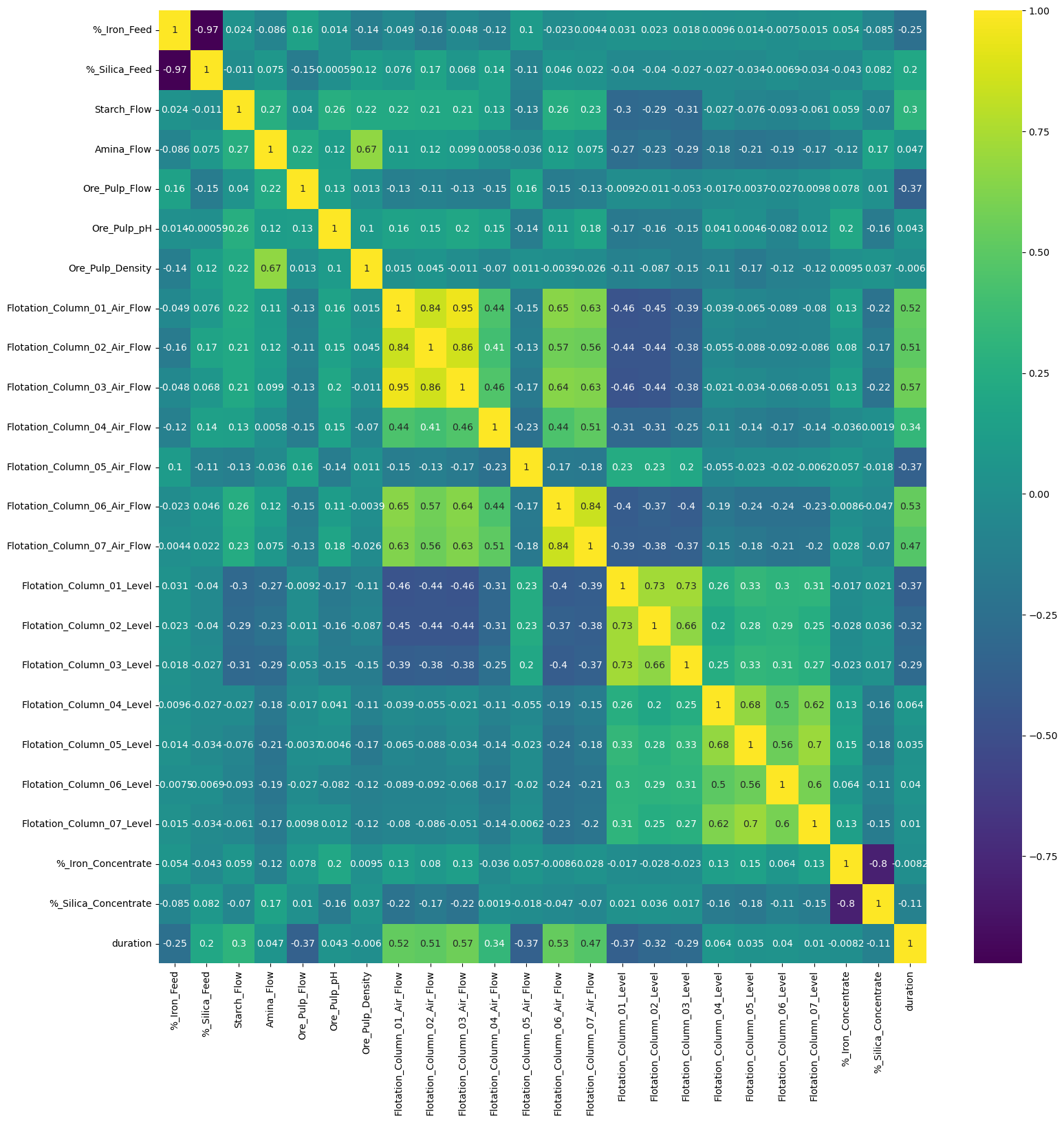
- First you can tell the % of silica is negatively corelated to the % of iron concentrate, which makes sense since they are separately desired and undesired substance of the process, purity and impurity
- Secondly, the airflow in column 1 is in general positively corelated to the airflow in column 2-7 except for column 5, which arise a potential doubt deserve diving deeper. Also we can tell the larger the airflow is, the lower the flotation level is in the responding column.
- Thirdly, we can see from the correlation heat map that the ore pulp flow and starch flow barely have influence on the final product – % iron and % silica. on top of the fact that they both have positive corelation with the airflow measured in the columns, we can later verify the underlying collinearity by eigenvalue.
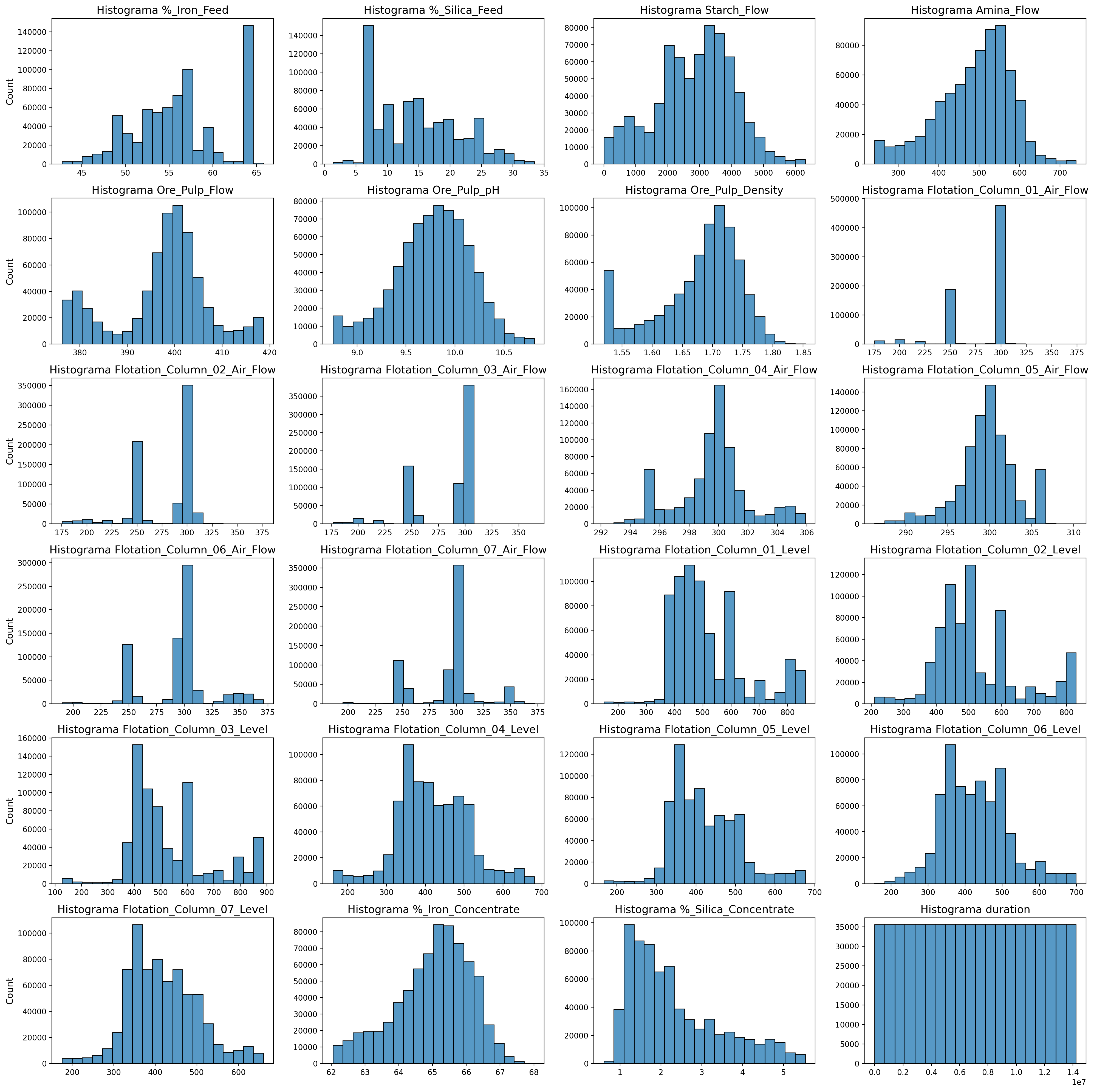
Feature Histogram-cross correlation and distribution
plt.figure(figsize=(20,20),dpi=200)
for i , n in enumerate(dt_df.columns.to_list()):
plt.subplot(6,4,i+1)
ax = sns.histplot(data=dt_df, x=n, kde=False, bins=20)#, multiple="stack")
plt.title(f"Histograma {n}", fontdict={"fontsize":14})
plt.xlabel("")
plt.ylabel(ax.get_ylabel(), fontdict={"fontsize":12})
if i not in [0,4,8,12,16,20,24]:
plt.ylabel("")
plt.tight_layout();
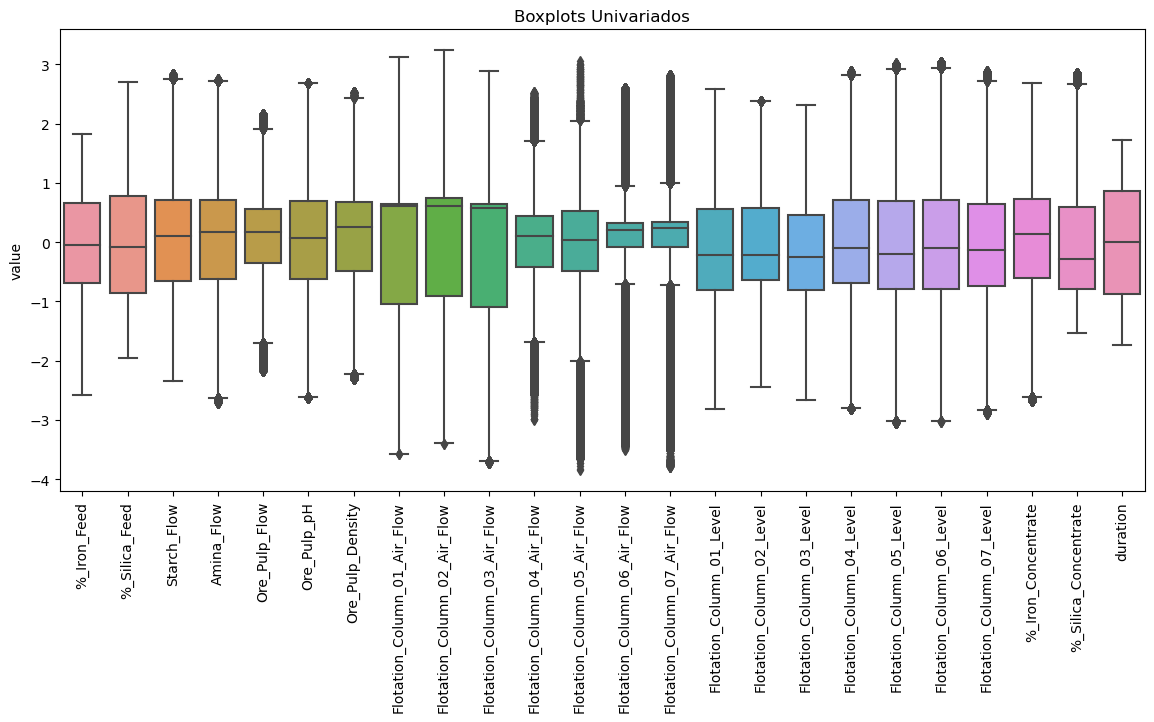
Feature Boxplot-distribution
scaler = StandardScaler()
z = pd.DataFrame(scaler.fit_transform(dt_df), columns=dt_df.columns, index=dt_df.index)
z = z.melt()
plt.figure(figsize=(14,6),dpi=100)
sns.boxplot(x=z["variable"], y=z["value"]);
plt.xticks(rotation=90);
plt.xlabel("");
plt.title("Boxplots Univariados");
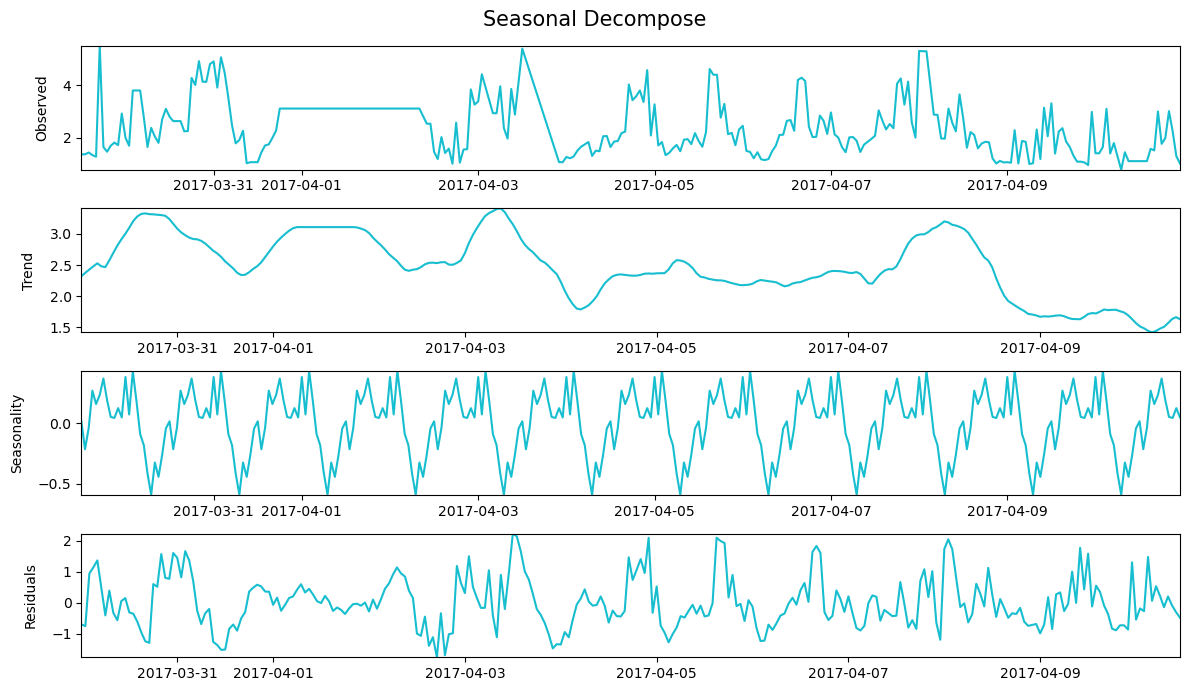
Timeseries lag trend
df_h = dt_df.resample('H').first()
df_h.index.names = ['Date']
result = seasonal_decompose(df_h["%_Silica_Concentrate"][:300])
def decompose_plot(result):
fig,axes = plt.subplots(ncols=1,nrows=4,figsize=(12,7))
axes[0].plot(result.observed.index, result.observed.values, color='tab:cyan')
axes[0].set_ylabel("Observed",fontdict={"size":10,},labelpad=10)
axes[0].set_xticks(axes[0].get_xticks(), rotation=45)
axes[1].plot(result.trend.index, result.trend.values, color='tab:cyan')
axes[1].set_ylabel("Trend",fontdict={"size":10,},labelpad=10)
axes[2].plot(result.seasonal.index, result.seasonal.values, color='tab:cyan')
axes[2].set_ylabel("Seasonality",fontdict={"size":10,},labelpad=10)
axes[3].plot(result.resid.index, result.resid.values, color='tab:cyan')
axes[3].set_ylabel("Residuals",fontdict={"size":10,},labelpad=10)
for n in range(0,4):
axes[n].autoscale(axis="both",tight=True)
fig.suptitle("Seasonal Decompose",fontsize=15)
plt.tight_layout()
decompose_plot(result)
Supervised TimeDiff Machine Learning
dt_df.columns
Index(['%_Iron_Feed', '%_Silica_Feed', 'Starch_Flow', 'Amina_Flow',
'Ore_Pulp_Flow', 'Ore_Pulp_pH', 'Ore_Pulp_Density',
'Flotation_Column_01_Air_Flow', 'Flotation_Column_02_Air_Flow',
'Flotation_Column_03_Air_Flow', 'Flotation_Column_04_Air_Flow',
'Flotation_Column_05_Air_Flow', 'Flotation_Column_06_Air_Flow',
'Flotation_Column_07_Air_Flow', 'Flotation_Column_01_Level',
'Flotation_Column_02_Level', 'Flotation_Column_03_Level',
'Flotation_Column_04_Level', 'Flotation_Column_05_Level',
'Flotation_Column_06_Level', 'Flotation_Column_07_Level',
'%_Iron_Concentrate', '%_Silica_Concentrate', 'duration'],
dtype='object')
list_cols = [col for col in dt_df.columns.to_list()] # Use this if I want to include all explanatory variables.
list_cols.remove("%_Silica_Concentrate")
list_cols.remove("%_Iron_Concentrate")
# Resample the original df every 15 minutes.
df_15 = dt_df.resample("15min").first()
df_15 = df_15.drop("%_Iron_Concentrate", axis=1)
window_size = 3
# Taking advantage of the time factor, I add lagged explanatory variables at 15min, 30min, and 45min, respectively
# (these are the ones that were updated every 20s)
for col_name in list_cols:
for i in range(window_size):
df_15[f"{col_name} ({-15*(i+1)}mins)"] = df_15[f"{col_name}"].shift(periods=i+1)
# Resample from 15min to 1hour
df_h = df_15.resample('H').first()
# Here I change the name of my index.
df_h.index.names = ['Date']
# Add lagged values of the target variable as explanatory variables, lagged by 1, 2, and 3 hours respectively.
col_name = "%_Silica_Concentrate"
window_size = 3
for i in range(window_size):
df_h[f"{col_name} ({-i-1}h)"] = df_h[f"{col_name}"].shift(periods=i+1)
# Make the target variable the first column.
x = df_h.columns.to_list()
x.remove("%_Silica_Concentrate")
x.insert(0, "%_Silica_Concentrate")
df_h = df_h[x]
df_h = df_h.dropna() # Remove rows with missing values due to the shift().
df_h = df_h.astype("float32") # Convert data to float32 for faster computation.
I create performance reporting functions for my model, a tracker to keep track of metrics, and also add a function to perform a time series split (where temporal order is respected).
Helper Functions
def plot_time_series(timesteps, values, start=0, end=None, label=None):#, #color="blue"):
"""
Plots timesteps (a series of points in time) against values (a series of values across timesteps).
Parameters
----------
timesteps : array of timestep values
values : array of values across time
format : style of plot, default "."
start : where to start the plot (setting a value will index from start of timesteps & values)
end : where to end the plot (similar to start but for the end)
label : label to show on plot about values, default None
"""
# Plot the series
sns.lineplot(x=timesteps[start:end], y=values[start:end], label=label)
plt.xlabel("Date")
plt.ylabel("% Silica Concentrate")
if label:
plt.legend(fontsize=14) # make label bigger
plt.grid(True)
def timeseries_report_model(y_test, model_preds, tracker_df="none", model_name="model_unknown", seasonality=1, naive=False):
mae = round(mean_absolute_error(y_test, model_preds), 4)
rmse = round(mean_squared_error(y_test, model_preds) ** 0.5, 4)
mase = round(mean_absolute_scaled_error(y_test, model_preds, seasonality, naive),4)
r2 = round(r2_score(y_test, model_preds), 4)
mape = round(mean_absolute_percentage_error(y_test, model_preds), 4)
print("MAE: ", mae)
print("RMSE :", rmse)
print("MASE :", mase)
print("R2 :", r2)
print("MAPE :", mape)
if isinstance(tracker_df, pd.core.frame.DataFrame):
tracker_df.loc[tracker_df.shape[0]] = [
model_name, mae, rmse, mase, r2, mape]
else:
pass
LSTM model
# Make the train/test splits
def make_train_test_splits(windows, labels, test_split=0.2):
"""
Splits matching pairs of winodws and labels into train and test splits.
"""
split_size = int(len(windows) * (1-test_split)) # this will default to 80% train/20% test
train_windows = windows[:split_size]
train_labels = labels[:split_size]
test_windows = windows[split_size:]
test_labels = labels[split_size:]
return train_windows, test_windows, train_labels, test_labels
df_forecast = df_h.copy()
df_forecast.shape
(3946, 92)
tracker = timeseries_models_tracker_df()
X = df_forecast.drop("%_Silica_Concentrate", axis=1)
y = df_forecast["%_Silica_Concentrate"]
train_windows, test_windows, train_labels, test_labels = make_train_test_splits(X,y,test_split=0.1)
len(train_windows), len(test_windows), len(train_labels), len(test_labels)
(3551, 395, 3551, 395)
warnings.filterwarnings("ignore")
def objective(trial, X,y):
param_grid = {
"random_state": 123,
"verbosity": -1,
"boosting_type": trial.suggest_categorical("boosting_type", ['gbdt']),#,"goss"]),
"device_type": trial.suggest_categorical("device_type", ['cpu']),
"n_estimators": trial.suggest_int("n_estimators", 25, 10000,step=100), #for large datasets this should be very high
"learning_rate": trial.suggest_float("learning_rate", 0.001, 0.3),
"num_leaves": trial.suggest_int("num_leaves", 10, 190), # Also change this for large datasets, should be small to avoid overfitting
"max_depth": trial.suggest_int("max_depth", 2, 80),
"min_child_samples": trial.suggest_int("min_child_samples", 10, 800, step=20), #modify this for large datasets, causes overfittin if too low
"reg_alpha": trial.suggest_float("reg_alpha", 0.0001, 1000, log=True),
"reg_lambda": trial.suggest_float("reg_lambda", 0.0001, 1000, log=True),
#"min_split_gain": trial.suggest_float("min_split_gain", 0, 3),
"subsample": trial.suggest_float("subsample", 0.05, 1, step=0.05),
"subsample_freq": trial.suggest_categorical("subsample_freq", [0,1]), #[0,1] bagging
"colsample_bytree": trial.suggest_float("colsample_bytree", 0.3, 1, step=0.1),
}
#min_data_in_leaf # min_child_samples
# RMSE (Root Mean Squared Error)
def rmse(y_val,y_pred):
is_higher_better = False
name = "rmse"
value = mean_squared_error(y_val,y_pred, squared=False)
return name, value, is_higher_better
cv_scores = np.empty(5)
tscv = TimeSeriesSplit(n_splits=5, max_train_size=None)
for idx , (train_index, val_index) in enumerate(tscv.split(X)):
X_train, X_val = X.iloc[train_index], X.iloc[val_index]
y_train, y_val = y.iloc[train_index], y.iloc[val_index]
model = LGBMRegressor(objective="regression", silent=True,**param_grid)
model.fit(
X_train,
y_train,
eval_set=[(X_val, y_val)],
eval_metric=rmse,
early_stopping_rounds=100,
categorical_feature="auto", # cat_idx, #specifiy categorial features.
callbacks=[LightGBMPruningCallback(trial, "l2", report_interval=20)], # Add a pruning callback
verbose=0)
preds = model.predict(X_val)
rmse = mean_squared_error(y_val,preds, squared=False)
cv_scores[idx] = rmse
return np.mean(cv_scores)
study = optuna.create_study(direction="minimize", study_name="LGBM Regressor",sampler=TPESampler(),
pruner=optuna.pruners.PercentilePruner(50, n_startup_trials=5, n_warmup_steps=50))
#study.enqueue_trial(try_this_first)
func = lambda trial: objective(trial, train_windows, train_labels)
study.optimize(func, n_trials=100)
[32m[I 2023-04-14 20:06:45,615][0m A new study created in memory with name: LGBM Regressor[0m
[32m[I 2023-04-14 20:06:46,696][0m Trial 0 finished with value: 0.8109004958352836 and parameters: {'boosting_type': 'gbdt', 'device_type': 'cpu', 'n_estimators': 8325, 'learning_rate': 0.2136801933659405, 'num_leaves': 122, 'max_depth': 24, 'min_child_samples': 290, 'reg_alpha': 0.4089594656291566, 'reg_lambda': 233.41962530839834, 'subsample': 0.55, 'subsample_freq': 1, 'colsample_bytree': 0.5}. Best is trial 0 with value: 0.8109004958352836.[0m
[32m[I 2023-04-14 20:06:47,790][0m Trial 1 finished with value: 0.8149075483133782 and parameters: {'boosting_type': 'gbdt', 'device_type': 'cpu', 'n_estimators': 2625, 'learning_rate': 0.20374443271553472, 'num_leaves': 24, 'max_depth': 65, 'min_child_samples': 550, 'reg_alpha': 0.00011716269934726968, 'reg_lambda': 0.005714841791728027, 'subsample': 0.45, 'subsample_freq': 0, 'colsample_bytree': 0.8}. Best is trial 0 with value: 0.8109004958352836.[0m
......
[32m[I 2023-04-14 20:08:36,711][0m Trial 99 finished with value: 0.718496606821031 and parameters: {'boosting_type': 'gbdt', 'device_type': 'cpu', 'n_estimators': 3425, 'learning_rate': 0.25476699329382246, 'num_leaves': 78, 'max_depth': 44, 'min_child_samples': 30, 'reg_alpha': 0.006442917820858156, 'reg_lambda': 0.9818132921273436, 'subsample': 0.45, 'subsample_freq': 1, 'colsample_bytree': 0.8}. Best is trial 71 with value: 0.7022349235029833.[0m
# Train again using the optimized hyperparameters
params = study.best_params
model = LGBMRegressor(objective="regression",random_state=123,**params)
model.fit(train_windows, train_labels)
LGBMRegressor(colsample_bytree=0.8, device_type='cpu',
learning_rate=0.2514458173401127, max_depth=48,
min_child_samples=90, n_estimators=2125, num_leaves=128,
objective='regression', random_state=123,
reg_alpha=0.0042343839479169615, reg_lambda=8.01369798587411,
subsample=0.6500000000000001, subsample_freq=1)
preds = model.predict(test_windows)
timeseries_report_model(test_labels, preds, tracker, model_name="Experiment , LightGBM",
seasonality=1, naive=False)
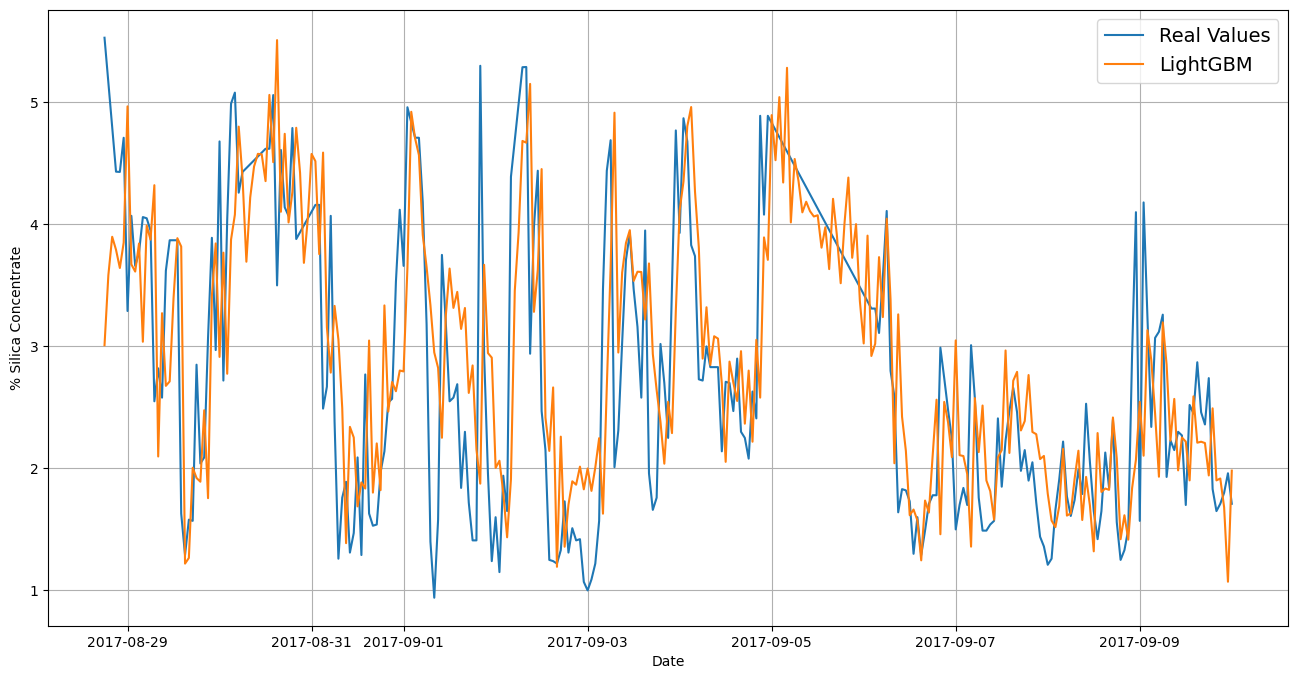
MAE: 0.6045
RMSE : 0.8177
MASE : 1.1928
R2 : 0.5233
MAPE : 0.2752
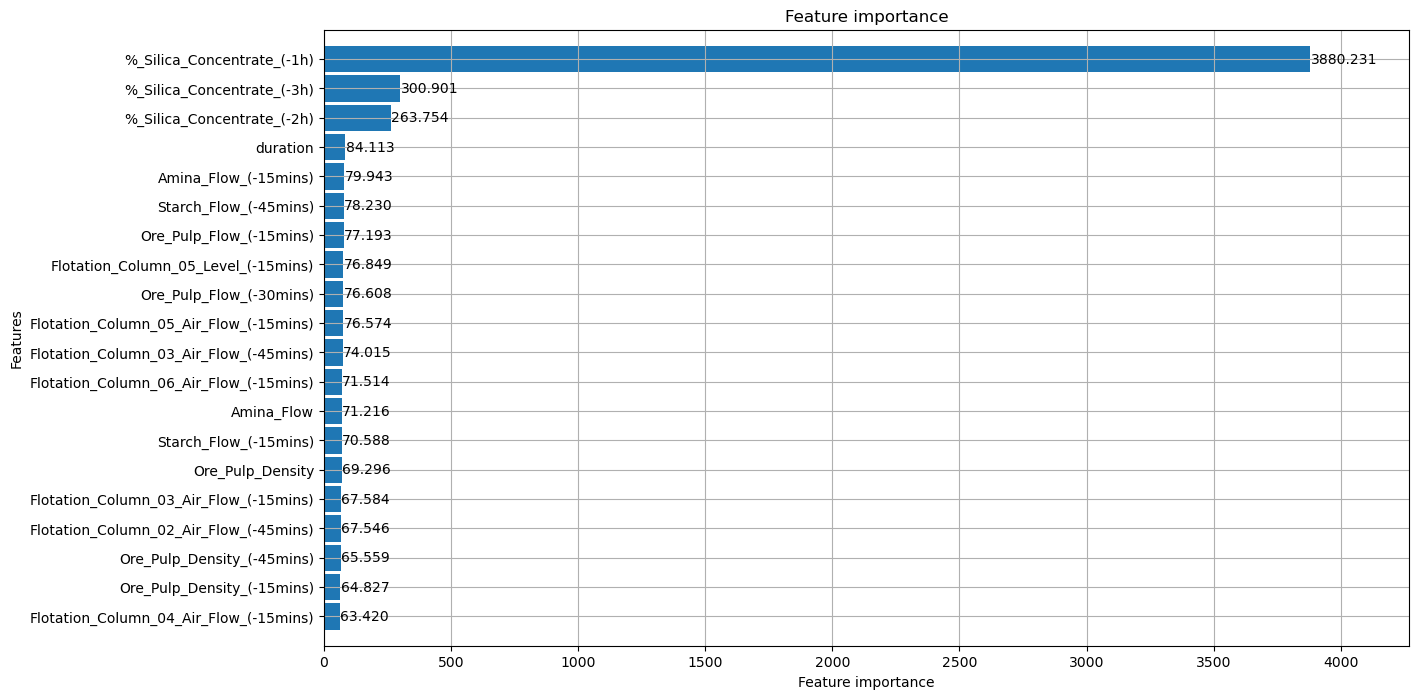
plot_importance(model, max_num_features = 20, height=.9, importance_type="gain", figsize=(14,8));
plt.figure(figsize=(16,8),dpi=100)
plot_time_series(test_labels.index, test_labels, label="Real Values", start=100)
plot_time_series(test_labels.index, preds,label="LightGBM", start=100)